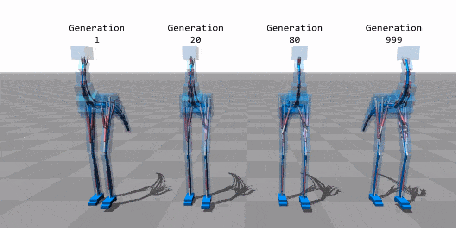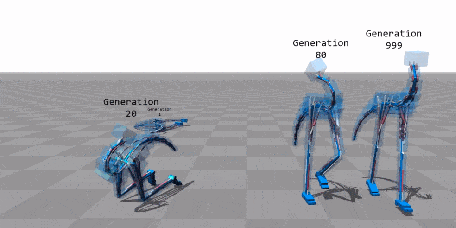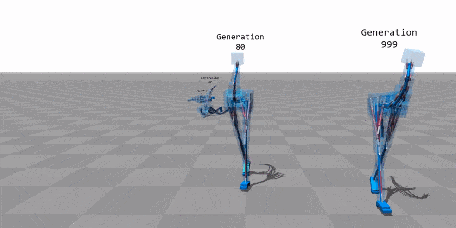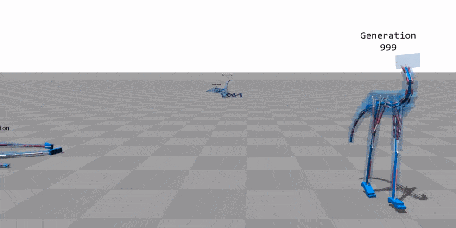
When Biology Meets Data Science—An Introduction to Neuroevolution¶
Tim Hargreaves
PyData Manchester
23rd June 2020
Introduction¶
About Me¶
- MathStat Undergraduate at University of Warwick
- Data Scientist at AstraZeneca
- Computational Statistics and Machine Learning
Format¶
- High-level with links to further reading
- Source code/presentation will be linked at the end
- Chance for Q&As after talk
Motivation¶
Conventional Methods for Designing/Tuning Neural Networks¶
| Method | Flaws |
|---|---|
| Copy someone else | Requires trust/might not be appropriate |
| Use intuition | No reproducibility; requires said intuition |
| Brute force/random search | Computational complexity |
| Gradient-based methods | Lack of convexity; non-differentiable hyperparameters |
| Bayesian methods | Deciding priors/measuring convergence can be difficult |
A Contrasting Approach¶
- Evolution offers a solution
- Can we implement something similar?
Evolutionary Algorithms¶
Evolution Recap¶
- An organism/individual is differentiated by its unique genetic code
- This genetic code will express itself in physical attributes
- These attributes impact the likelihood of survival
- Surviving organisms are more likely to reproduce
- Reproduction allows an organism to partially pass on their genes
- Mutations can occur at any time to change an organism's genes
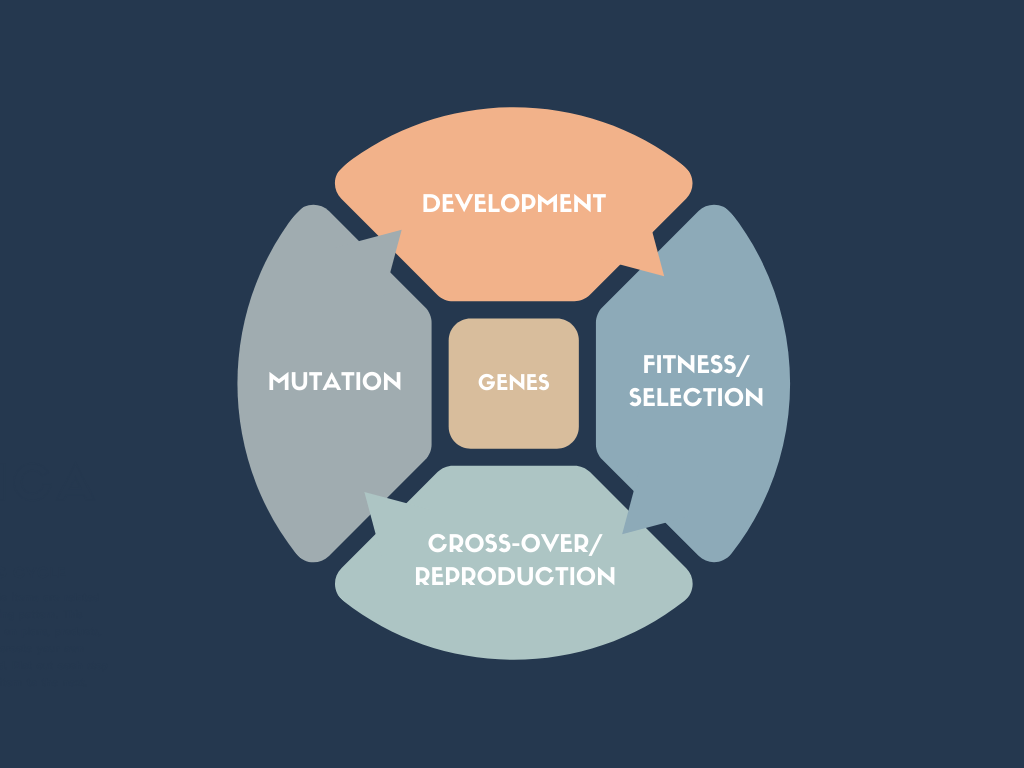
Components of Evolution¶
- Genes
- Fitness/Selection
- Cross-over/Reproduction
- Mutation
Genes¶
- The smallest unit of selection
- Possible expressions of a particular gene are called alleles
- Contributes to the physical attributes of the organism
Fitness/Selection¶
- Each organism has an abstract value of fitness
- This relates to its chance of survival/reproduction
- Genes that encourage fitness are more likely to disseminate
Cross-over/Reproduction¶
- Two organisms can reproduce to have children
- Each child will take half their genetic material from each parent
- This loosely corresponds to 'mixing' the genes of the parents
Mutation¶
- Genes can mutate to change their expression
- This can occur at any time but is more likely at birth
Bringing it Together¶
Application to Optimisation¶
- Genes -> Parameters
- Fitness -> Score
- Definitions of reproduction/mutation
- No one 'correct' formulation
Example: Quadratic Regression¶
Task¶
- Find optimal (least squares) coefficents to fit noisy quadratic data
Genes¶
gene_pool = {
'a': (-3, 3),
'b': (-3, 3),
'c': (-3, 3),
}
# Initialisation
genome = {
gene: random.uniform(alleles[0], alleles[1])
for gene, alleles in population.gene_pool.items()
}
Fitness/Selection¶
self._fitness = -np.mean(np.square(y_hat - y))
ranked_organisms = sorted(self.organisms,
key=lambda org: org.fitness,
reverse=True)
survived = ranked_organisms[:self.retain_length]
for org in ranked_organisms[self.retain_length:]:
if len(survived) >= self.size:
break
if self.random_select > random.random():
survived.append(org)
# Determine how many children to create
shortfall = self.size - len(survived)
children = []
while len(children) < shortfall:
mother = random.choice(survived)
father = random.choice([s for s in survived
if s is not mother])
children.extend(
Organism.breed(mother, father,
min(2, shortfall - len(children)))
)
self.organisms = survived + children
Cross-over/Reproduction¶
for gene in mother.population.gene_pool.keys():
# Convex combination of parents' alleles
p = random.random()
genome[gene] = p * mother.genome[gene] + \
(1-p) * father.genome[gene]
Mutation¶
for gene, alleles in self.population.gene_pool.items():
if self.population.mutation_chance > random.random():
# Convex combination of current allele and random
# choice from the gene pool
p = (1 + random.random()) / 2
self.genome[gene] = p * self.genome[gene] + \
(1-p) * random.uniform(alleles[0], alleles[1])
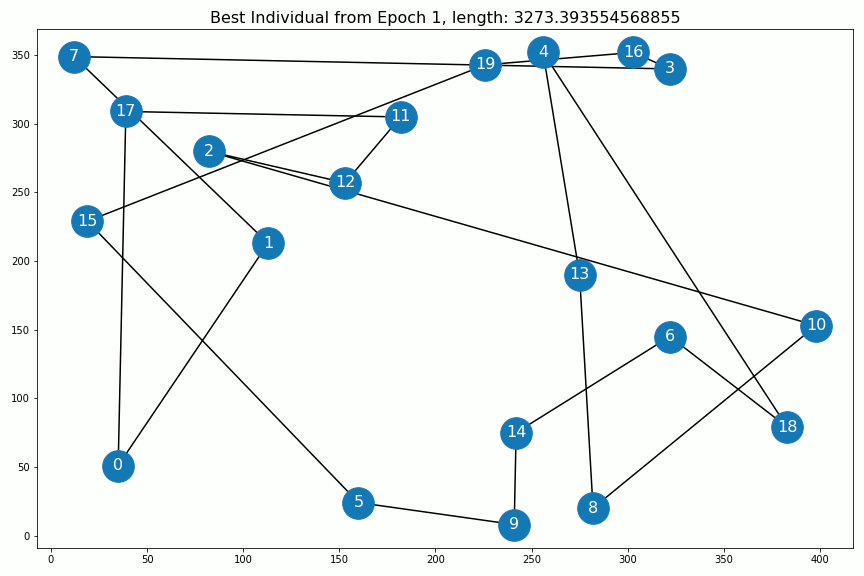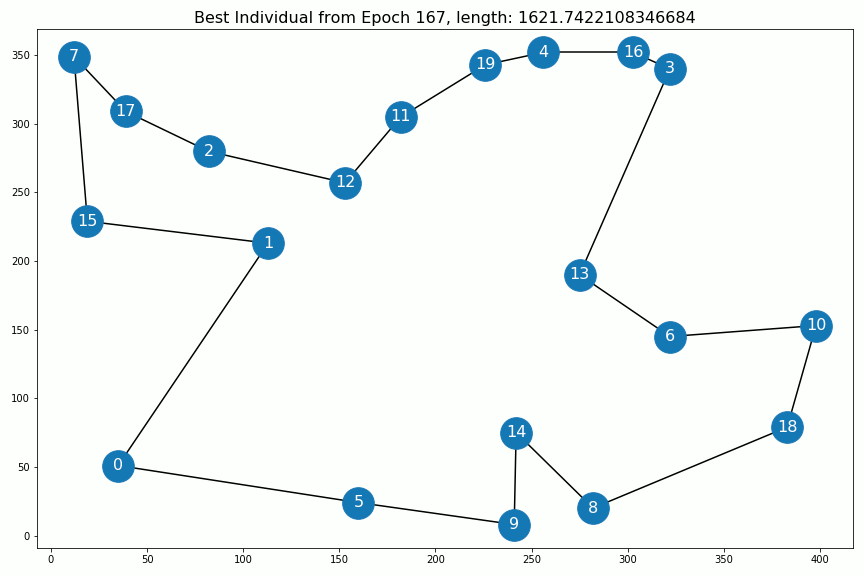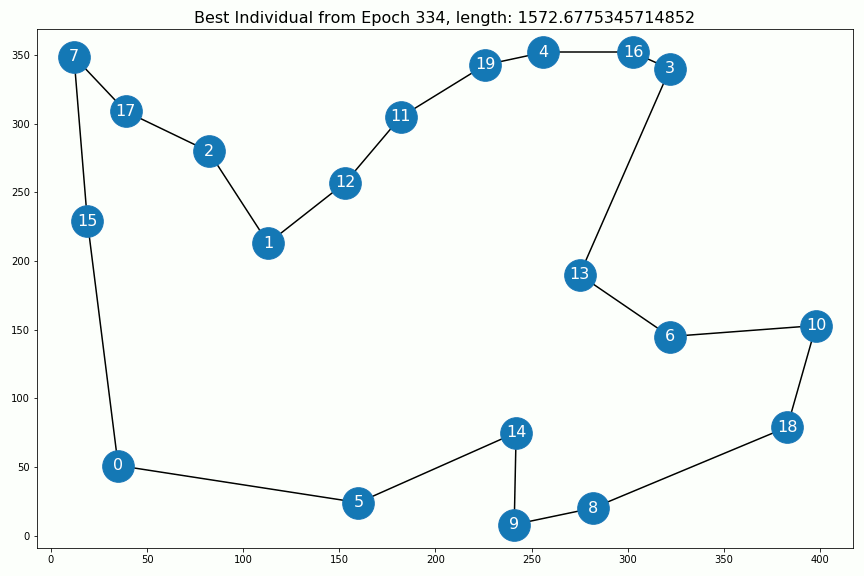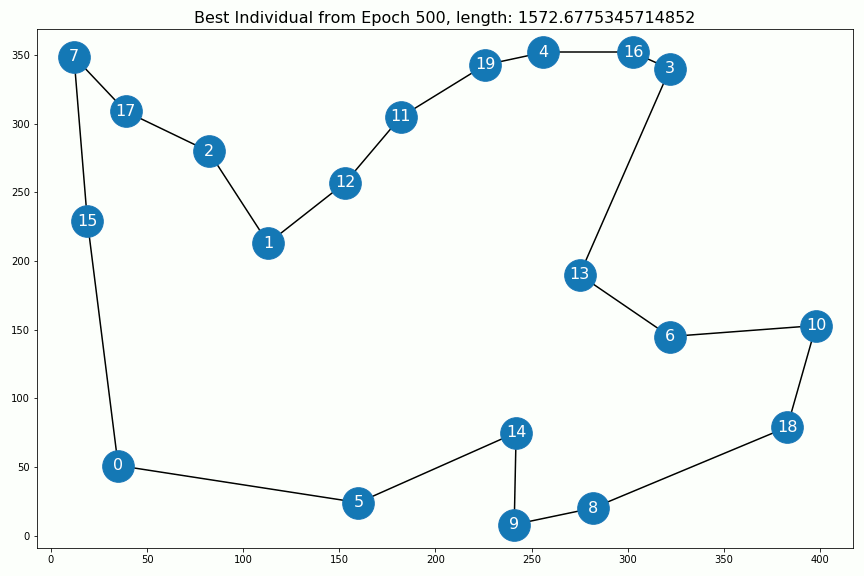
Results¶
Out[5]:
Out[6]:
Comparison to Grid Search¶
- Grid search on integer lattice: $7^3=343$ combinations
- For precision of $0.5$ this increases to $13^3=2197$
- Evolutionary method above had $20$ organisms per generation
- Linear in number of generations
- Reasonable convergence at $5$ generations
Comparison to Gradient Descent¶
$$
\begin{align}
\sum\left(y-\hat{y}'\right)^2 &:= \sum\left(y-\hat{y} + c - \hat{c}\right)^2 \\ &= \sum\left[\left(y-\hat{y}\right)^2 + \left(y-\hat{y}\right)\left(c - \hat{c}\right) + \left(c-\hat{c}\right)^2\right] \\ &= \sum\left(y-\hat{y}\right)^2 + K \qquad \left[\sum\left(y-\hat{y}\right)=0\right]
\end{align}$$
Key point: How easy is it to calculate gradients?
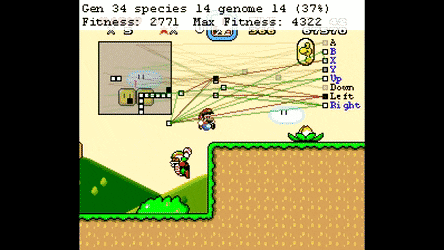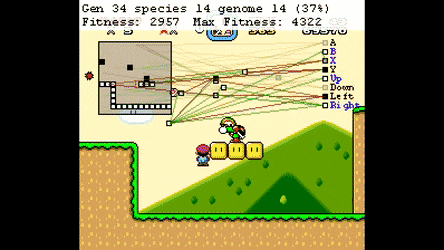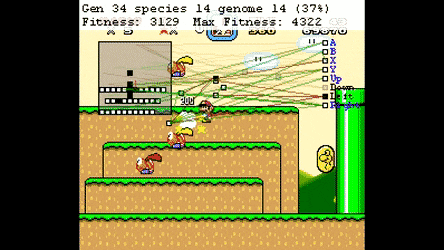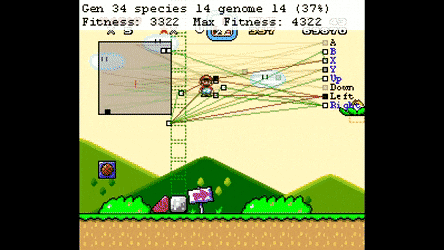
Evolutionary Algorthms in the Wild¶
Neural Networks¶
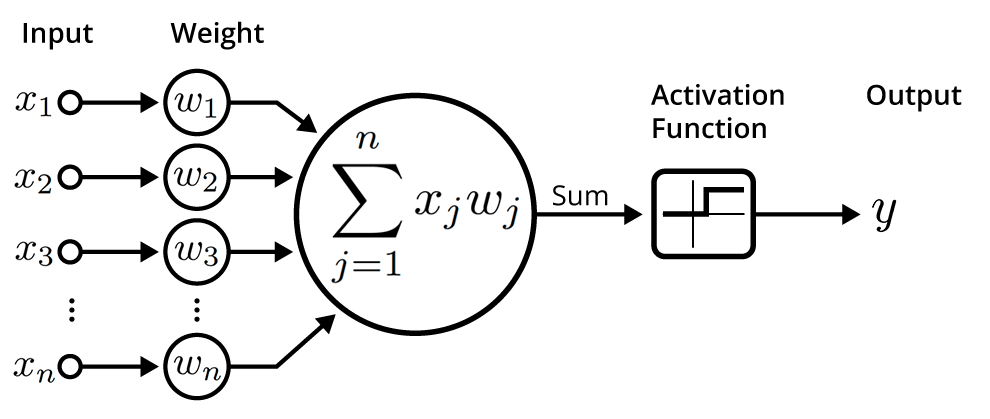
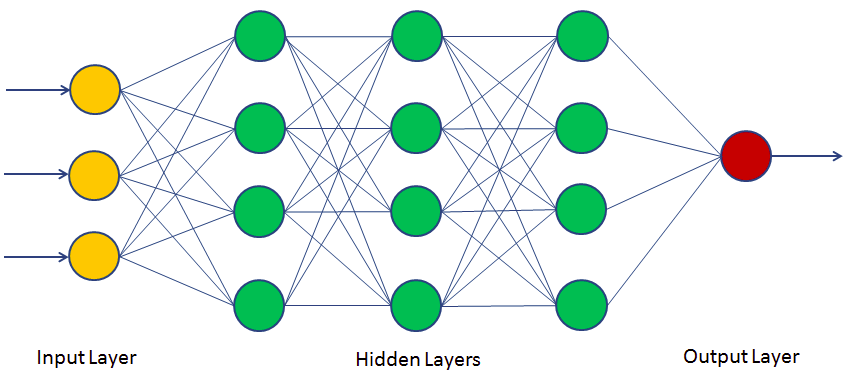
Neural Network Recap¶
- Loosely modelled on the architecture of the brain
- Capture complex interations between inputs
Hyperparameters¶
- Learning rate?
- Batch size?
- Weight/bias initialisation?
- Which optimiser?
Neuroevolution¶
Optimisation Space¶
- Number of layers
- Number of neurons per layer
- Whether to include bias
- Whether to use dropout/how much
- Activation types
- Optimiser
Search space size: ~10000 combinations
Genes¶
gene_pool = {
'num_neurons': [32, 64, 128, 256, 512, 768, 1024],
'num_layers': [1, 2, 3, 4, 5],
'use_bias': [True, False],
'dropout_rate': [0., .1, .2, .3, .4],
'activation': ['relu', 'elu', 'tanh', 'sigmoid'],
'optimiser': ['rmsprop', 'adam', 'sgd', 'adagrad',
'adadelta', 'adamax', 'nadam'],
}
# Initialisation
genome = {
gene: random.choice(alleles)
for gene, alleles in population.gene_pool.items()
}
Fitness/Selection¶
self._fitness = self.train_and_evaluate()
def train_and_evaluate(self):
"""Train this organism's model and report the final accuarcy."""
if self.population.verbose:
print(f"Training model {self.id}")
# For prototype, only train for one epoch
self.model.fit(X_train, y_train,
batch_size=1024,
epochs=1,
verbose=False,
validation_data=(X_test, y_test))
score = self.model.evaluate(X_test, y_test, verbose=False)
return score[1] # accuracy
ranked_organisms = sorted(self.organisms,
key=lambda org: org.fitness,
reverse=True)
survived = ranked_organisms[:self.retain_length]
for org in ranked_organisms[self.retain_length:]:
if len(survived) >= self.size:
break
if self.random_select > random.random():
survived.append(org)
# Determine how many children to create
shortfall = self.size - len(survived)
children = []
while len(children) < shortfall:
mother = random.choice(survived)
father = random.choice([s for s in survived
if s is not mother])
children.extend(
Organism.breed(mother, father,
min(2, shortfall - len(children)))
)
self.organisms = survived + children
Cross-over/Reproduction¶
for gene in mother.population.gene_pool.keys():
genome[gene] = random.choice((
mother.genome[gene], father.genome[gene]
))
Mutation¶
for gene, alleles in self.population.gene_pool.items():
if self.population.mutation_chance > random.random():
self.genome[gene] = random.choice(alleles)
Results¶
Out[16]:
num_neurons: 512 num_layers: 5 use_bias: False dropout_rate: 0.1 activation: relu optimiser: adam
Comparison to Random Search¶
Randomly created 25 models without improvement Randomly created 50 models without improvement Randomly created 75 models without improvement Randomly created 100 models without improvement Randomly created 125 models without improvement Randomly created 150 models without improvement Randomly created 175 models without improvement Randomly created 200 models without improvement Randomly created 225 models without improvement Randomly created 250 models without improvement Randomly created 275 models without improvement Randomly created 300 models without improvement Randomly created 325 models without improvement Randomly created 350 models without improvement Randomly created 375 models without improvement Randomly created 400 models without improvement Randomly created 425 models without improvement Randomly created 450 models without improvement Randomly created 475 models without improvement Randomly created 500 models without improvement Giving up...
Conclusion¶
Where next?¶
- Productionising solution
- Experimenting with different selection/reproduction/mutation techniques
- Parallel training on multiple GPUs
- More sophisticated initialisation
Wrapping Up¶
- Source material/presentation: https://github.com/THargreaves/neuroevolution-talk
- Thank you for listening
- Any questions?